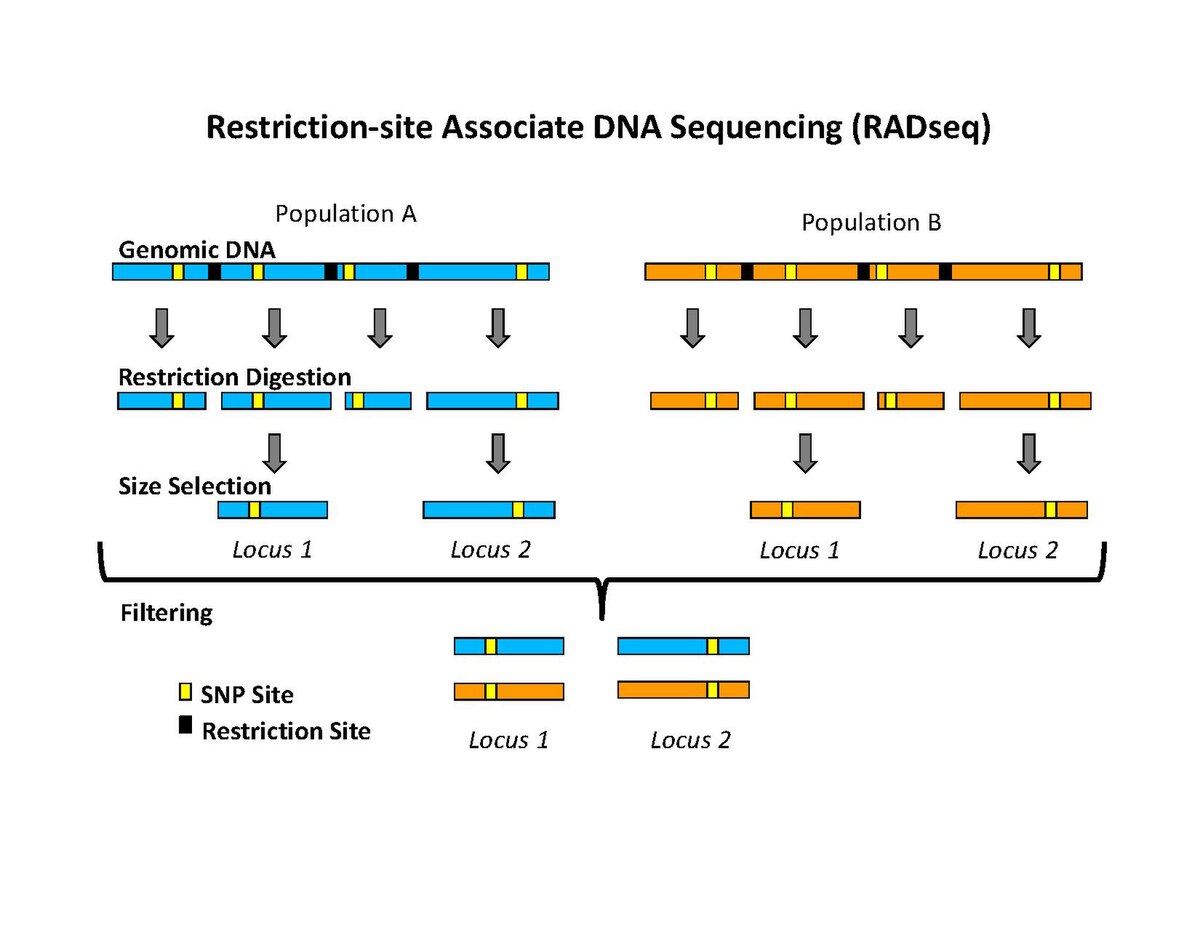
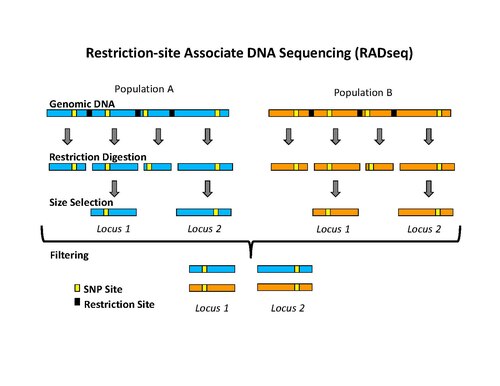
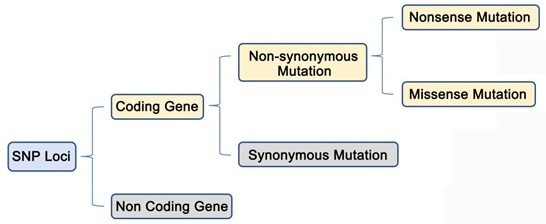
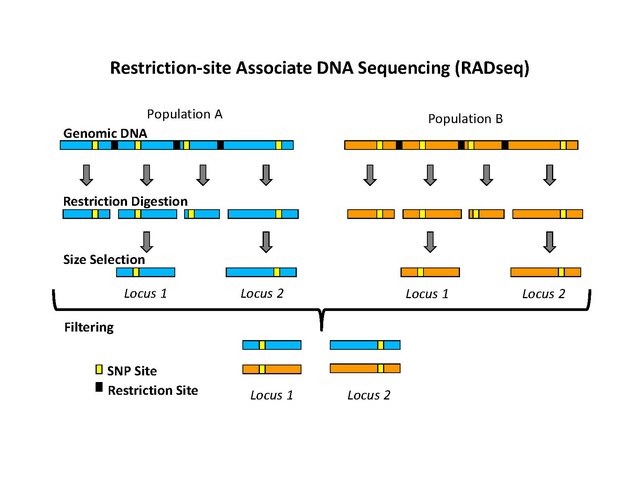
Analysis of genome-wide SNPs based on 2b-RAD sequencing of pooled samples reveals signature of selection in different populations of Haemonchus contortus | SpringerLink
2b-RAD genotyping for population genomic studies of Chagas disease vectors: Rhodnius ecuadoriensis in Ecuador | PLOS Neglected Tropical Diseases

CD Genomics on Twitter: "The Advantages and Workflow of 2b-RAD We reviewed the main advantages of 2b-RAD over other RAD sequencing technologies. https://t.co/yYkGdqbKeI https://t.co/X9eWfPUOVY" / Twitter

An improved 2b-RAD approach (I2b-RAD) offering genotyping tested by a rice (Oryza sativa L.) F2 population – topic of research paper in Biological sciences. Download scholarly article PDF and read for free

Would an RRS by any other name sound as RAD? - Campbell - 2018 - Methods in Ecology and Evolution - Wiley Online Library

Protocol used for ENU treatment and estimation of mutation spectrum by... | Download Scientific Diagram

Screening and characterization of sex-specific markers for the hybrid (Megalobrama amblycephala ♀ × Ancherythroculter nigrocauda ♂) based on 2b- RAD and transcriptome sequencing - ScienceDirect

Figure 1 from Sequence Capture versus Restriction Site Associated DNA Sequencing for Shallow Systematics. | Semantic Scholar

Brage NMBU: Genetics of resistance to photobacteriosis in gilthead sea bream (Sparus aurata) using 2b-RAD sequencing
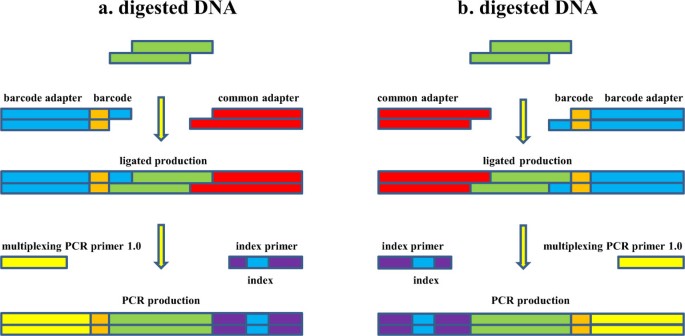
An improved 2b-RAD approach (I2b-RAD) offering genotyping tested by a rice (Oryza sativa L.) F2 population | BMC Genomics | Full Text

Agronomy | Free Full-Text | Optimizing ddRADseq in Non-Model Species: A Case Study in Eucalyptus dunnii Maiden
2b-RAD genotyping for population genomic studies of Chagas disease vectors: Rhodnius ecuadoriensis in Ecuador | PLOS Neglected Tropical Diseases
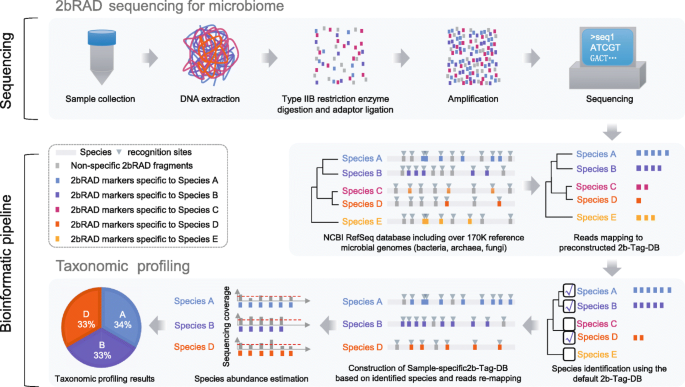
Species-resolved sequencing of low-biomass or degraded microbiomes using 2bRAD-M | Genome Biology | Full Text

Trade‐offs and utility of alternative RADseq methods: Reply to Puritz et al. - Andrews - 2014 - Molecular Ecology - Wiley Online Library

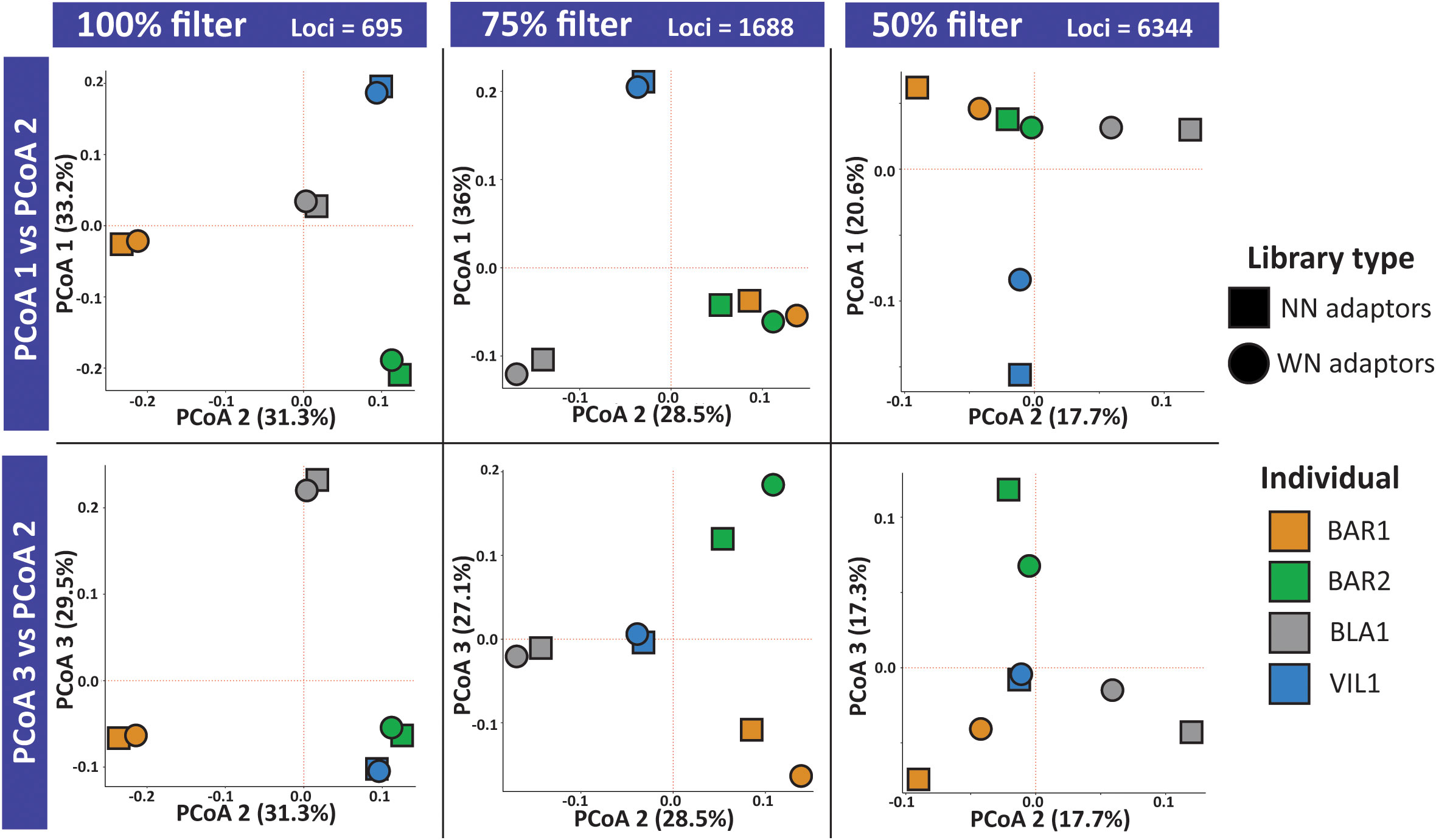

![ezRAD: a simplified method for genomic genotyping in non-model organisms [PeerJ] ezRAD: a simplified method for genomic genotyping in non-model organisms [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2013/203/1/fig-1-full.png)